Please visit JAWS Homepage for detailed documentation.
AAWS, GCNet, IMAU, POLENET, PROMICE, SCAR, NSIDC
Number of stations = 620
Number of station-years of data = 4000+
Size of data for all stations = ~4 GB
Earliest year of available data = 1931 (Station 040030 from NSIDC)
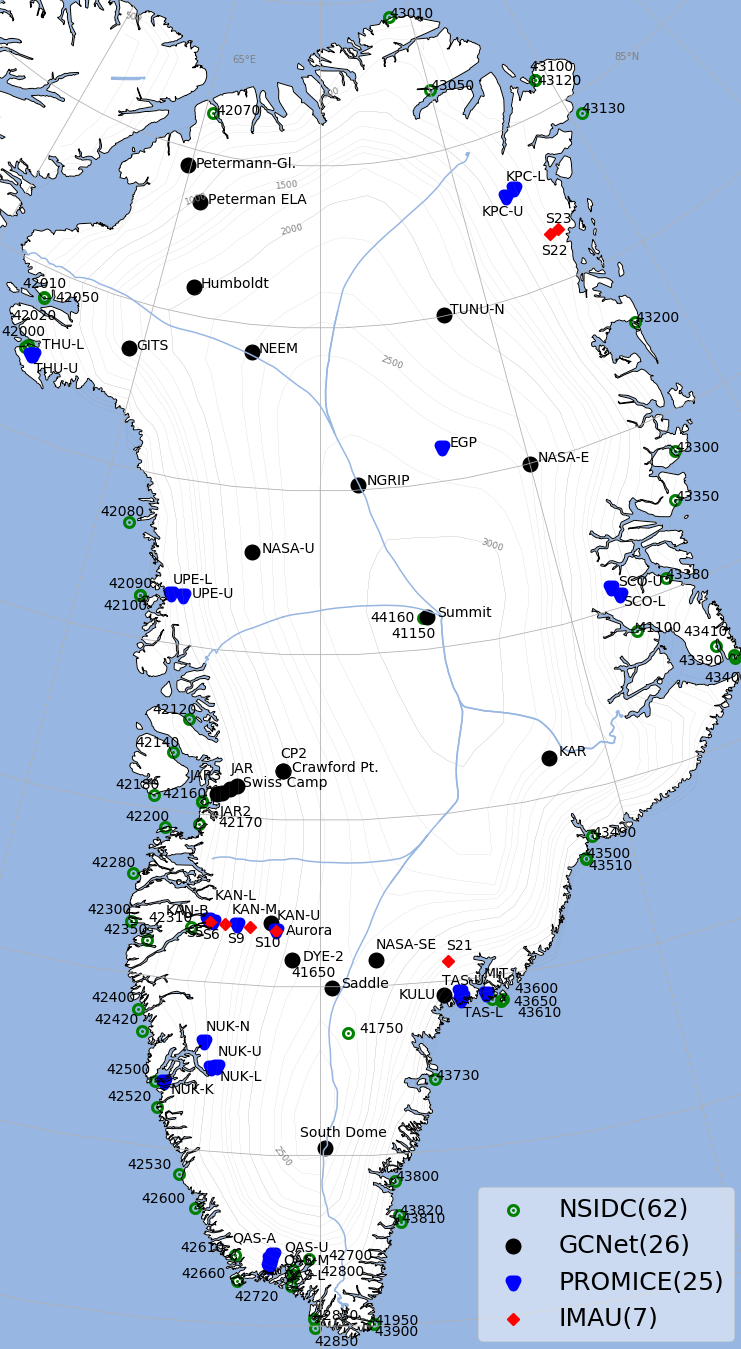
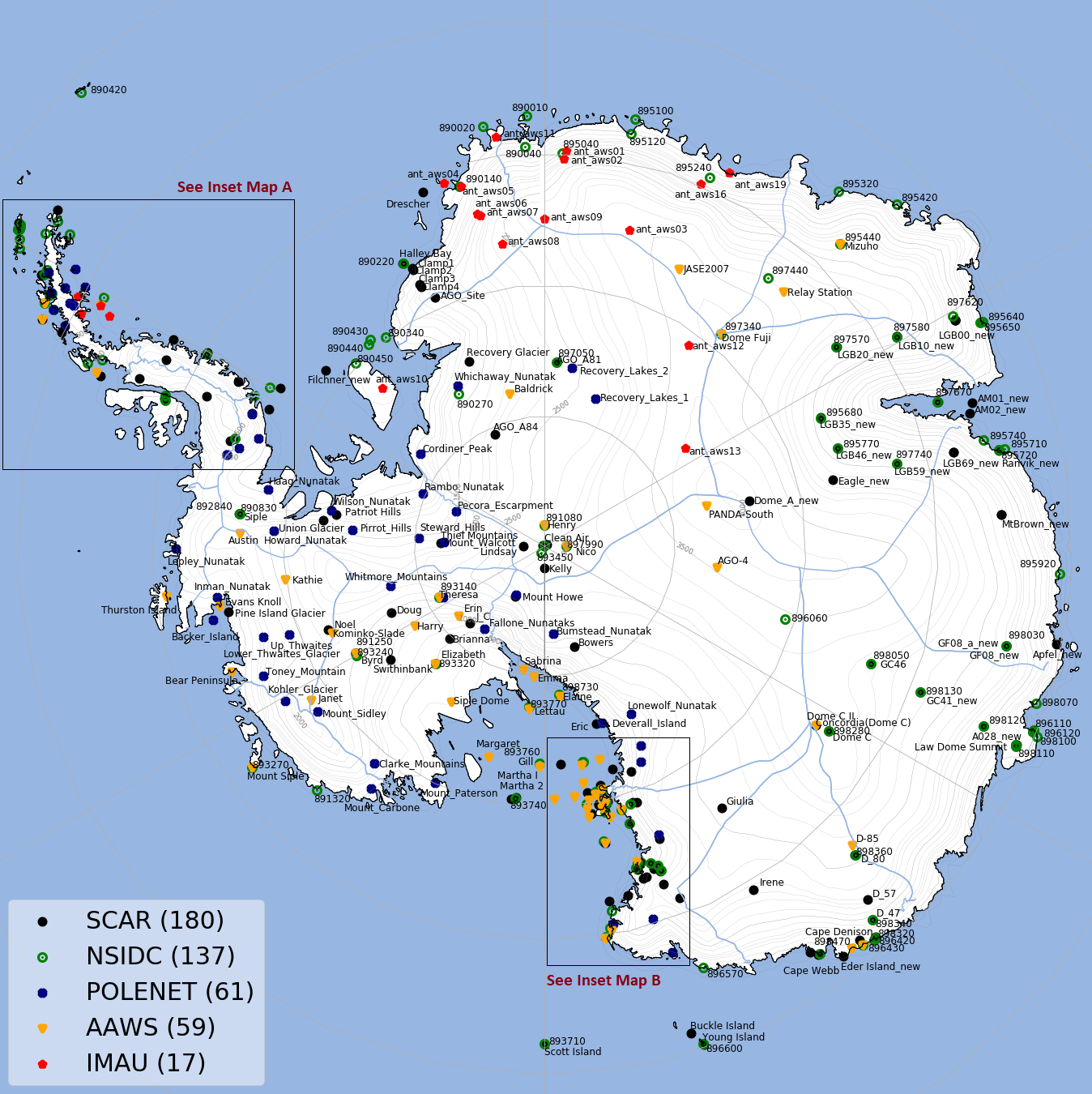
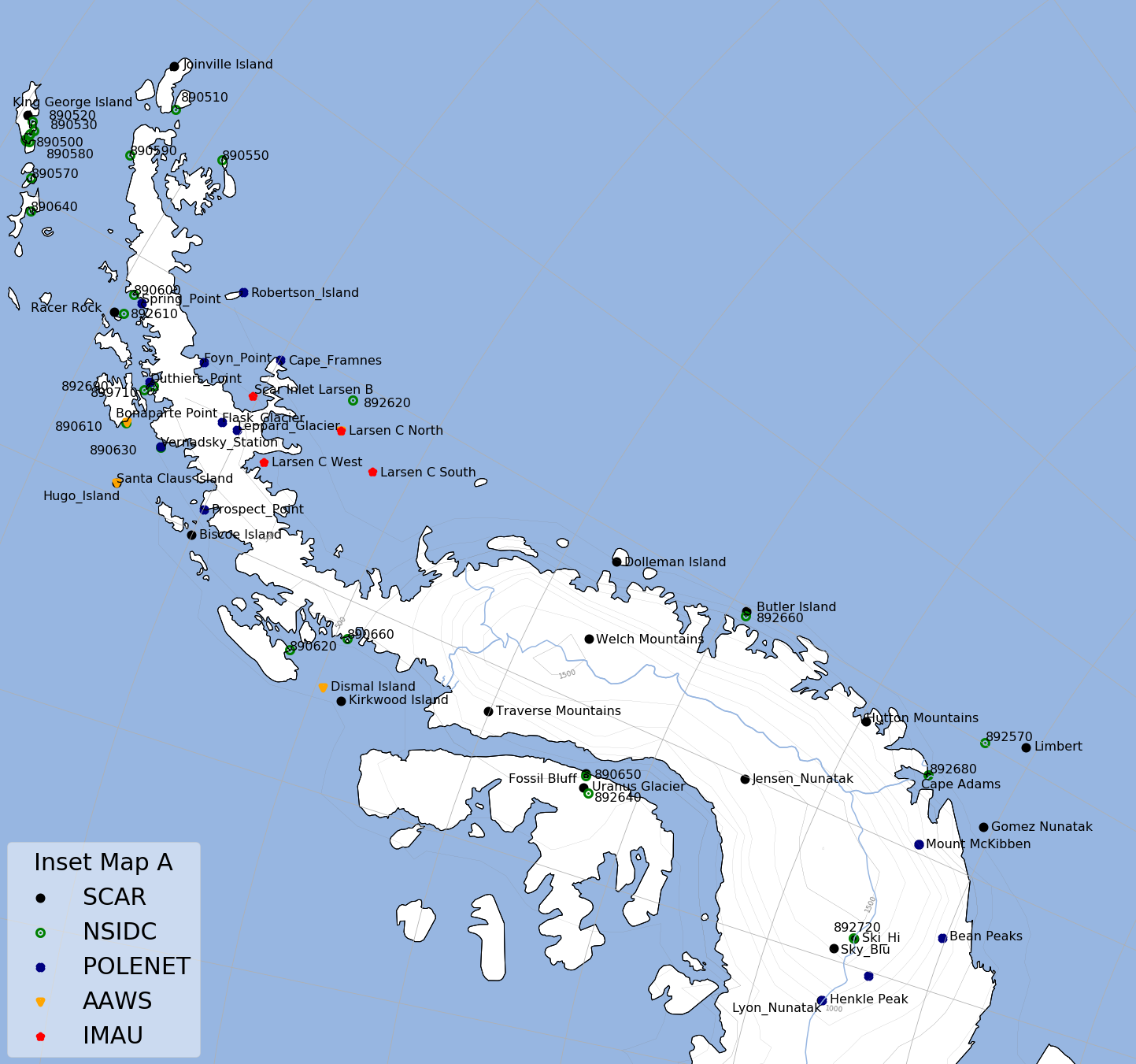
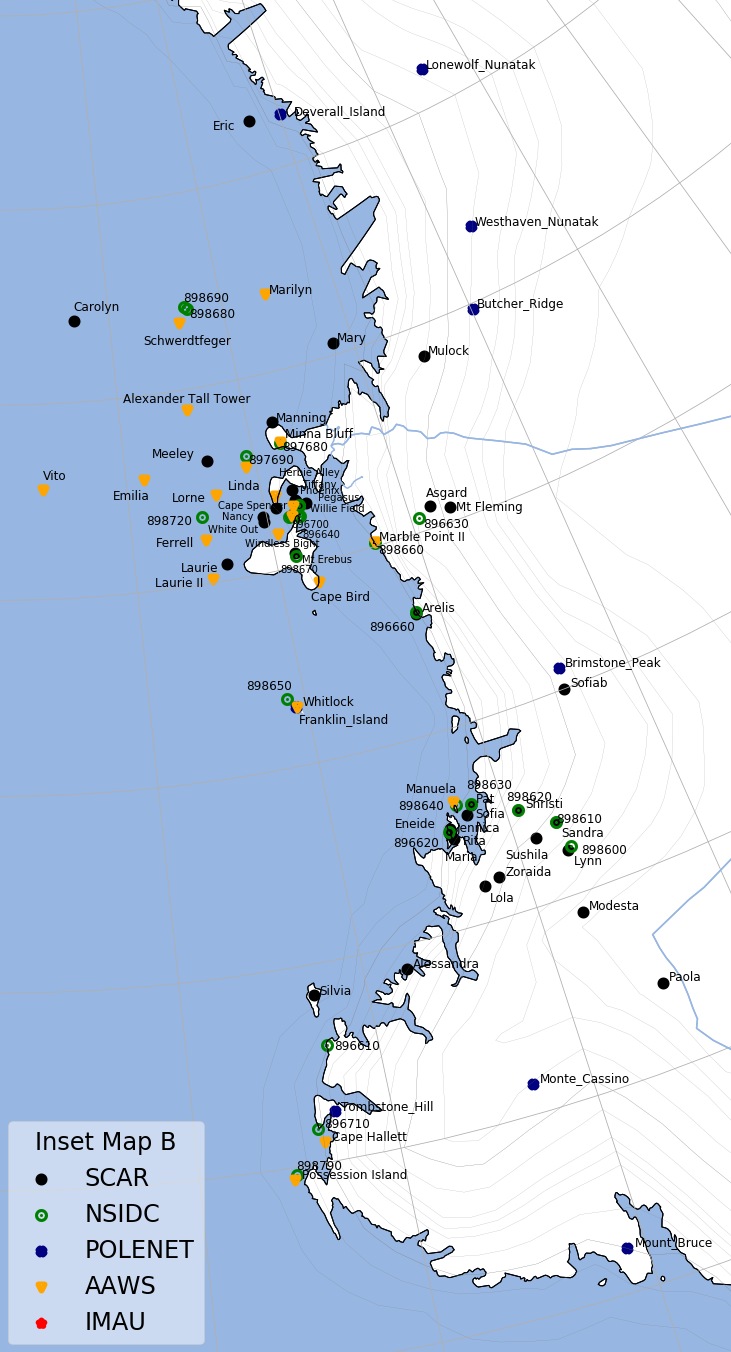
Maps of Antarctica and Greenland with location of all stations are in Stations Map section.
If your network is not in the above list and you would like it to be supported by JAWS,
please open an issue or contact Charlie Zender at zender@uci.edu
2019/09/30: Version 1.0 released: Stable, long-term release
2019/08/14: Version 0.9.3 released: Correct time for NSIDC files prior to 1970
2019/08/11: Version 0.9.1 released: Add new network NSIDC
2019/07/17: Version 0.9 released: Correct time variable for SCAR; Add hints
2019/05/16: Version 0.8.5 released: PROMICE v03 files supported; --hPa option
2019/05/03: Version 0.8.4 released: Analysis for txt files
2019/04/23: Version 0.8.3 released: Gradient fluxes for GCNet
2019/04/14: Version 0.8.2 released: Celsius option
2019/04/04: Version 0.8.1 released: RIGB fix 1 hr shift issue; CMIP naming
2019/03/30: Version 0.8 released: RIGB improvement, Documentation
2019/02/06: Version 0.7 released: POLENET network added
2019/01/11: Version 0.6.5 released: RIGB post-processing
2018/12/07: Version 0.6.3 released: RIGB adjusted fluxes archived
2018/10/31: Version 0.6 released: RIGB tilt correction
2018/10/08: Version 0.5 released: SCAR stations convertible to netCDF by JAWS
2018/05/23: Version 0.4 released: Pip installable, analysis.py callable from 'jaws' keyword
2018/04/18: Version 0.3 released: Conda installable
2018/01/22: Version 0.2 released: Conversion of GCNet, PROMICE and AAWS networks complete
2017/10/23: Version 0.1 released: Original scripts from Wenshan
JAWS is a scientific software workflow to ingest Level 2 (L2) data in the multiple formats now distributed, harmonize it into a common format, and deliver value-added Level 3 (L3) output suitable for distribution by the network operator, analysis by the researcher, and curation by the data center. NASA has funded JAWS (project summary) from 20171001–20190930.
Automated Weather Station (AWS) and AWS-like networks are the primary source of surface-level meteorological data in remote polar regions. These networks have developed organically and independently, and deliver data to researchers in idiosyncratic ASCII formats that hinder automated processing and intercomparison among networks. Moreover, station tilt causes significant biases in polar AWS measurements of radiation and wind direction. Researchers, network operators, and data centers would benefit from AWS-like data in a common format, amenable to automated analysis, and adjusted for known biases.
The immediate target recipient elements are polar AWS network managers, users, and data distributors. L2 borehole data suffers from similar interoperability issues, as does non-polar AWS data. Hence our L3 format will be extensible to global AWS and permafrost networks. JAWS will increase in situ data accessibility and utility, and enable new derived products.
Convert L2 data (usually ASCII tables) into a netCDF-based L3 format compliant with metadata conventions (Climate-Forecast and ACDD) that promote automated discovery and analysis.
Include value-added L3 features like the Retrospective, Iterative, Geometry-Based (RIGB) tilt angle and direction corrections, solar zenith angle, standardized quality flags, GPS-derived ice velocity, and turbulent fluxes.
Provide a scriptable API to extend the initial L2-to-L3 conversion to newer AWS-like networks and instruments.
- Python 2.7, 3.6, or 3.7 (as of JAWS version 0.7)
By far the simplest and recommended way to install JAWS is using conda (which is the wonderful package manager that comes with Anaconda or Miniconda distribution).
You can install JAWS and all its dependencies with:
$ conda install -c conda-forge jawsIf you do not use conda, you can install JAWS from source with:
$ pip install jaws(which will download the latest stable release from the PyPI repository and trigger the build process.)
pip defaults to installing Python packages to a system directory (such as /usr/local/lib/python2.7). This requires root access.
If you don't have root/administrative access, you can install JAWS using:
$ pip install jaws --user--user makes pip install packages in your home directory instead, which doesn't require any special privileges.
Users should periodically update JAWS to the latest version using:
$ conda update -c conda-forge jawsor
$ pip install jaws --upgradeJAWS is a command-line tool. Linux/Unix users can run JAWS from terminal and Windows users from Anaconda Prompt.
The current version can translate L2 ASCII data from the following networks to netCDF format:
- Antarctic Automatic Weather Stations (AAWS): Sample raw file can be downloaded from here. Right click on the link and select "Save link as".
- Greenland Climate Network (GCNet): Sample raw file can be downloaded from here
- Institute for Marine and Atmospheric Research (IMAU): Sample raw file for Antarctic stations can be downloaded from here and for Greenland stations can be downloaded from here
- Programme for Monitoring of the Greenland Ice Sheet (PROMICE): Sample raw file can be downloaded from here
- Scientific Committee on Antarctic Research (SCAR): Sample raw file can be downloaded from here
- The Polar Earth Observing Network (POLENET): Sample raw file can be downloaded from here
- National Snow & Ice Data Center (NSIDC): Sample raw file can be downloaded from here
Note:
For PROMICE, input file name must contain station name. e.g. 'PROMICE_KAN-B.txt' or 'KAN-B.txt' or 'Kangerlussuaq-B_abc.txt', etc.
For IMAU, input file name must start with network type(i.e. 'ant' or 'grl'), followed by a underscore and then station number. e.g. 'ant_aws01.txt' or 'ant_aws15_123.txt' or 'grl_aws21abc.txt', etc.
For SCAR, input file name must end with '_aws.dat'
For POLENET, input file name must start with 'polenet_'
The user provides the input file path. By default, the output file will be stored within the current directory with same name as of input file (e.g. PROMICE_EGP_20160501.nc). The user can optionally give their own output path/name. Execute this to get output file in current directory:
$ jaws ~/Downloads/PROMICE_EGP_20160501.txtor by specifying longer paths, and with options:
$ jaws -4 -o ~/Desktop/PROMICE_EGP_20160501.nc ~/Downloads/PROMICE_EGP_20160501.txt
$ jaws -4 -o ~/Desktop/GCNet_Summit_20140601.nc ~/Downloads/GCNet_Summit_20140601.txt
$ jaws -4 -o ~/Desktop/AAWS_AGO-4_20161130.nc ~/Downloads/AAWS_AGO-4_20161130.txtwhere the argument to the optional -o is the user-defined output filename
A list of all options can be found in here.
Download sample data from here
To run RIGB:
$ jaws ~/Downloads/gcnet_summit_20120817.txt --rigbRIGB uses climlab package to simulate clear-sky radiation.
In addition to input variables, JAWS provides following variables in output netCDF file to make data more useful:
- time (seconds since 1970-01-01 00:00:00)
- time_bounds
- sza (solar zenith angle)
- latitude
- longitude
- ice_gps_velocity_x, ice_gps_velocity_y, ice_gps_velocity_total (For stations that archive GPS position)
- year, month, day, hour
- adjusted_fsds (corrected downwelling shortwave flux)
Currently, the input file for analysis should be in netCDF format. So, first the raw ASCII files should be converted to netCDF using previous steps. We are working to make it accept ASCII files as input.
In the following examples we have used GCNet station at Summit, if you are using a separate network, you need to change the variable name accordingly.
JAWS has ability to analyse the data in multiple ways such as:
i. Diurnal:
JAWS can be used to plot monthly diurnal cycle to see hourly changes for any variable throughout the month.
The user needs to provide input file path, variable name (on which analysis needs to be done) and
analysis type (i.e. diurnal, monthly, annual or seasonal). The argument for analysis is -a, --anl or --analysis and
variable name is -v, --var or --variable.
We will take two examples here:
-
Case 1: The input file contains only 1-day data. We will consider the file converted previously i.e. GCNet_Summit_20120817.nc
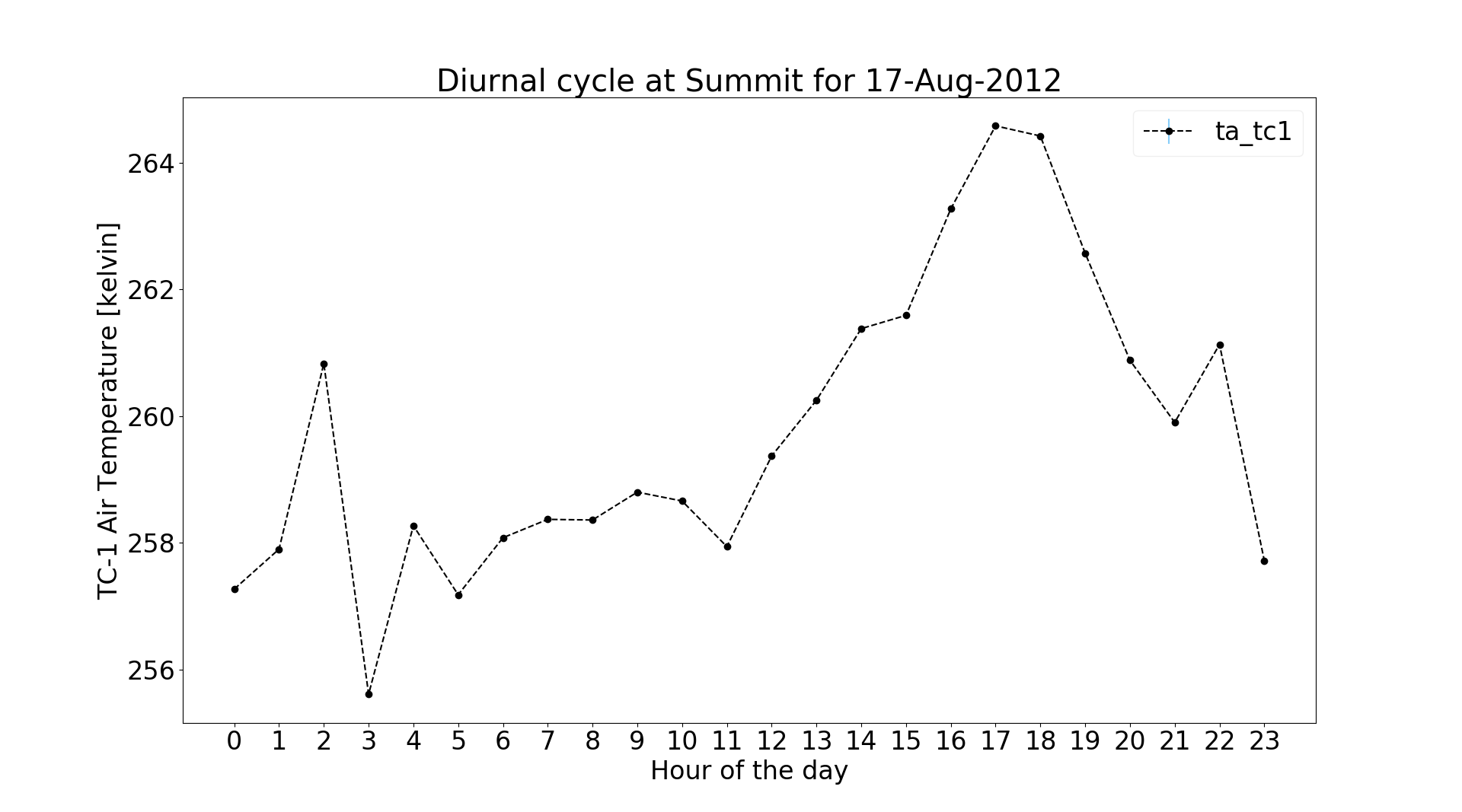
Use the following command to see how temperature varies throughout the day:
$ jaws -a diurnal -v ta_tc1 GCNet_Summit_20120817.nc -
Case 2: We will be using multi-year data from GCNet-Summit. We don't have permission to host this data.
Since, there are many years and months in this file, we need to provide for which year and month we want to do the analysis. The argument for year is
-y, --anl_yr or --analysis_yearand month is-m, --anl_mth or --analysis_month.If the input file contains data for only single year, then the user doesn't need to provide the '-y' argument. Similar is the case for '-m' (month) argument.
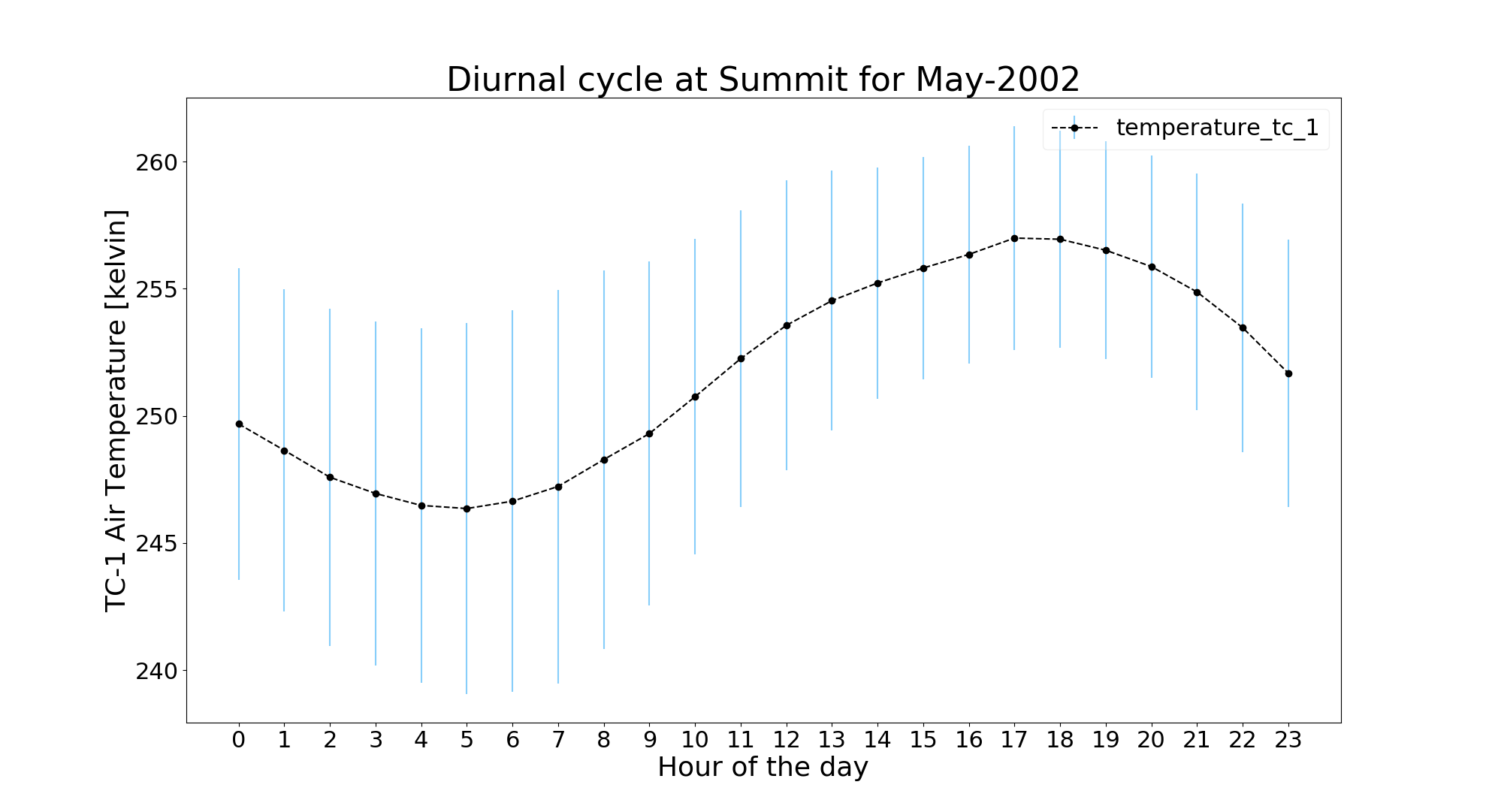
We will do the analysis for May-2002 at GCNet_Summit:
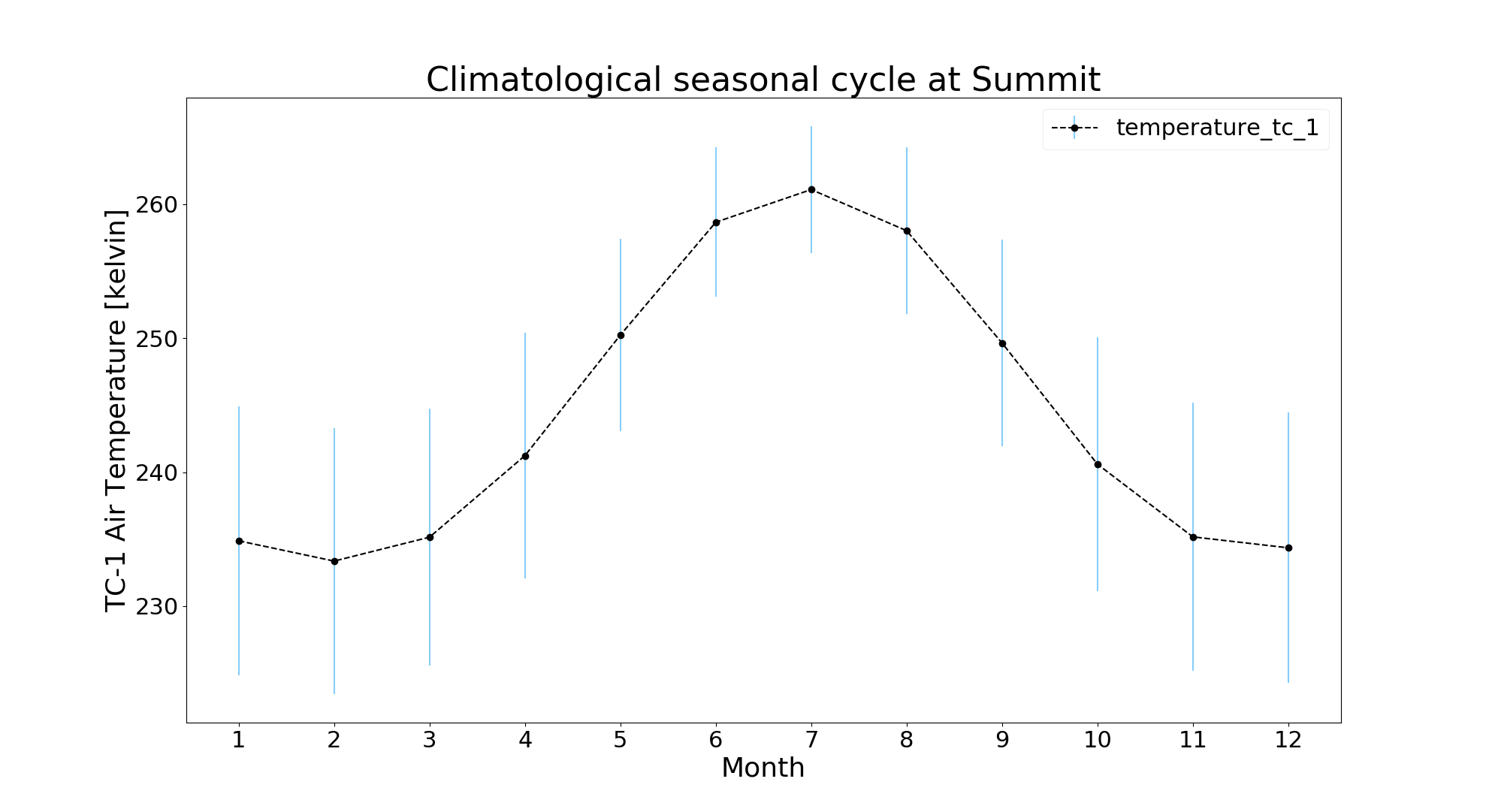
$ jaws -a diurnal -v ta_tc1 -y 2002 -m 5 gcnet_summit.ncThe blue error bar shows standard deviation for that hour across the month.
Important: This same file from Case 2 will be used for next three analysis because we need at-least monthly, yearly and
multi-yearly data respectively for them.
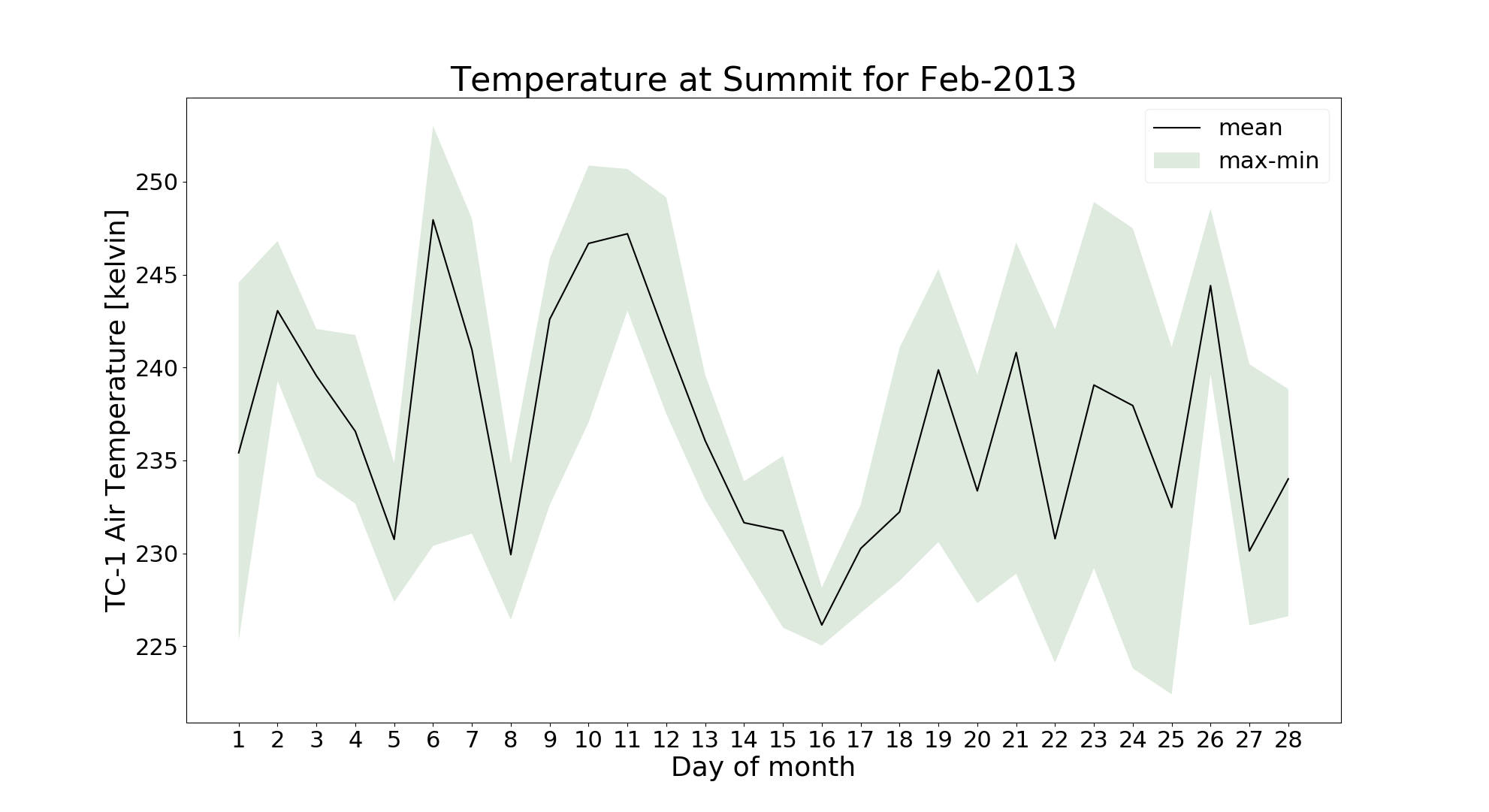
ii. Monthly:
In this analysis, we can analyze avg, max and min values for each day of a month for any variable
This time we will do it for temperature from different sensor for Feb-2013 as following:
$ jaws --anl monthly --var ta_cs1 --anl_yr 2013 --anl_mth 2 gcnet_summit.nc
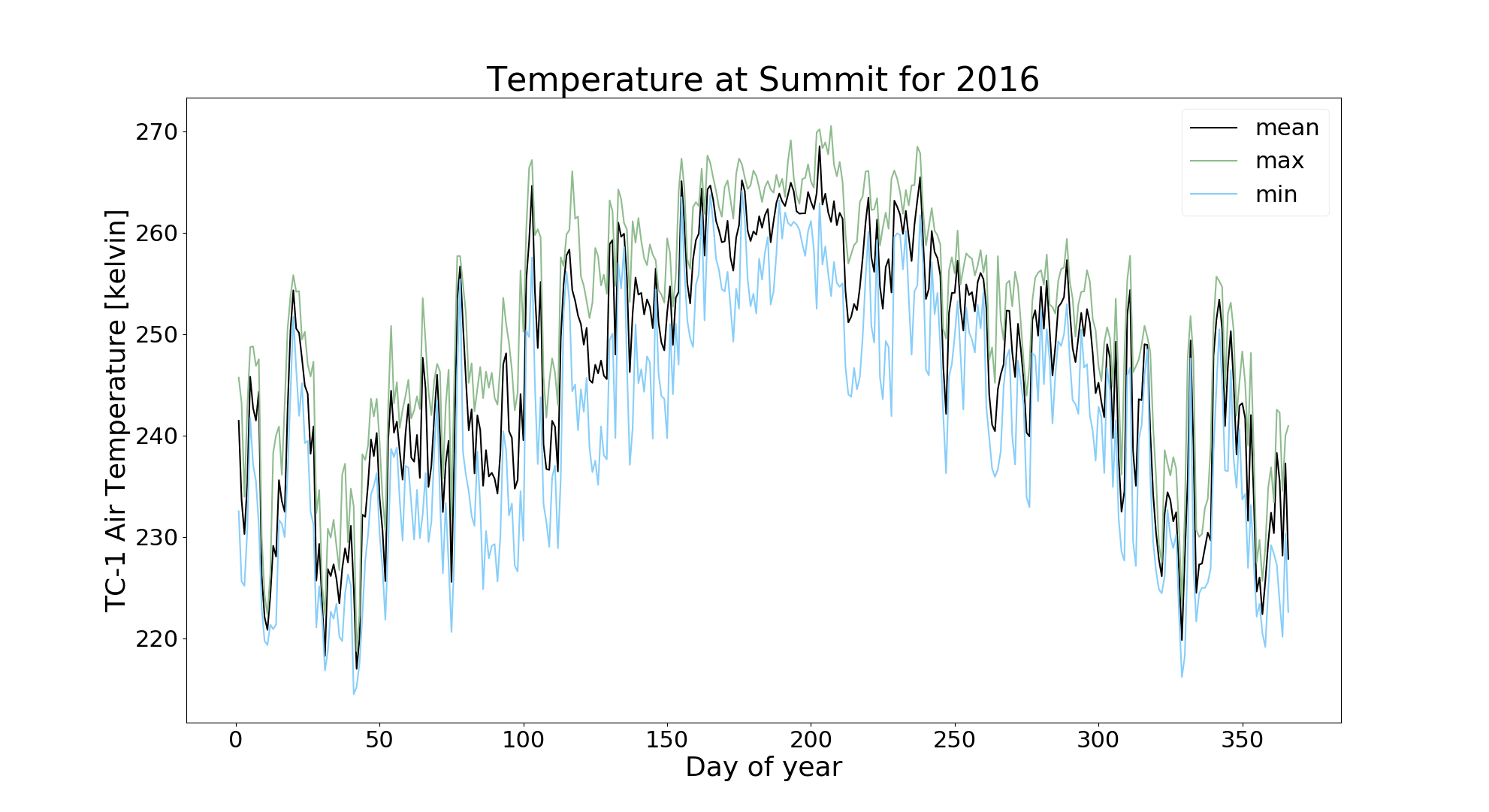
iii. Annual:
To plot an annual cycle with daily mean, max and min:
$ jaws --analysis annual --variable ta_tc1 --analysis_year 2016 gcnet_summit.nc
Note: Since it is annual plot, user shouldn't provide the '-m' argument
iv. Seasonal
Climatological seasonal cycle showing variation for each month through multiple years:
$ jaws -a seasonal -v ta_tc1 gcnet_summit.nc
Note: Since it is seasonal plot, user shouldn't provide both '-y', '-m' argument.
As of version 0.9, it takes about 3.5 minutes to process Summit(GCNet) data from 19960512 to 20170524
This software is being developed by the University of California Irvine under NASA Advanced Information Systems Technology (AIST) Proposal and Project 80NSSC17K0540.
For bugs, questions and discussions please use the GitHub Issues.
Copyright (C) 2017--2018 Regents of the University of California. You may redistribute and/or modify JAWS under the terms of the Apache License, Version 2.0.